 Image credit: Unsplash
Image credit: Unsplash
Abstract
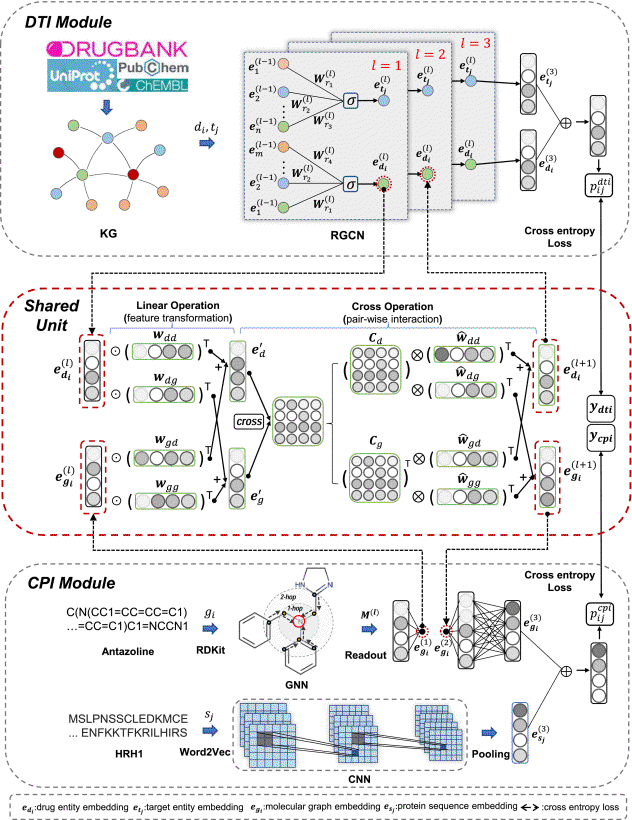
Molecular interaction prediction is essential in various applications including drug discovery and material science. The problem becomes quite challenging when the interaction is represented by unmapped relationships in molecular networks, namely molecular interaction, because it easily suffers from (i) insufficient labeled data with many false-positive samples, and (ii) ignoring a large number of biological entities with rich information in the knowledge graph. Most of the existing methods cannot properly exploit the information of knowledge graph and molecule graph simultaneously. In this paper, we propose a large-scale K nowledge G raph enhanced M ulti- T ask L earning model, namely KG-MTL, which extracts the features from both knowledge graph and molecular graph in a synergistic way. Moreover, we design an effective Shared Unit that helps the model to jointly preserve the semantic relations of drug entity and the neighbor structures of the compound in both knowledge graph and molecular graph. Extensive experiments on four real-world datasets demonstrate that our proposed KG-MTL outperforms the state-of-the-art methods on two representative molecular interaction prediction tasks drug-target interaction prediction and compound-protein interaction prediction. The source code of KG-MTL is available at https://github.com/xzenglab/KG-MTL.